Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Kataoka, Takahiro*; Shuto, Hina*; Naoe, Shota*; Yano, Junki*; Kanzaki, Norie; Sakoda, Akihiro; Tanaka, Hiroshi; Hanamoto, Katsumi*; Mitsunobu, Fumihiro*; Terato, Hiroaki*; et al.
Journal of Radiation Research (Internet), 62(5), p.861 - 867, 2021/09
Times Cited Count:5 Percentile:53.7(Biology)Pinak, M.; O'Neill, P.*; Fujimoto, Hirofumi; Nemoto, Toshiyuki*
AIP Conference Proceedings 708, p.310 - 313, 2004/05
The multiple nanosecond molecular dynamics simulations of DNA mutagenic oxidative lesion - 7,8-dihydro-8-oxoguanine (8-oxoG), complexed with the repair enzyme - human oxoguanine glycosylase 1 (hOGG1) in a physiological aqueous environment, were performed in order to describe the structural and energy changes in DNA and the dynamical process of DNA-enzyme complex formation. In complex the N-terminus of arginine 324 was found located close to the phosphodiester bond of the nucleotide with 8-oxoG enabling chemical reaction(s) between the amino acid and the lesion. The recognition of lesion on DNA, its recognition by repair enzyme and the formation of stable DNA-enzyme complex are necessary conditions for the onset of the successful enzymatic repair process.
Yokoya, Akinari
Hoshako, 17(3), p.111 - 117, 2004/05
To reveal the mechanism of oxidative base damages, such as 8-oxo-G, in DNA molecule by ionizing radiation, DNA base radicals were examined around oxygen and nitrogen K-edge region using an EPR spectrometer installed in a synchrotron soft X-ray beamline (BL23SU) in SPring-8. In situ measurements of EPR spectrum of guanine base revealed that short-lived transient radical species are specifically induced by photoexcitation of a 1s electron to 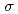 * antibonding orbital at carbonyl oxygen atom. They promptly disappear by "beam-off". On the other hand, a long-lived radical whose EPR spectrum is consistent with previous reports for guanine cation radical was accumulated during the irradiation. The yield of the stable radical decreased by These results indicate that chemically stable DNA base lesions, such as 8-oxo-G, would result from transient species that are inferred to be one electron oxidized radicals after decay of Auger final state.
* antibonding orbital at carbonyl oxygen atom. They promptly disappear by "beam-off". On the other hand, a long-lived radical whose EPR spectrum is consistent with previous reports for guanine cation radical was accumulated during the irradiation. The yield of the stable radical decreased by These results indicate that chemically stable DNA base lesions, such as 8-oxo-G, would result from transient species that are inferred to be one electron oxidized radicals after decay of Auger final state.
Pinak, M.
Computational Biology and Chemistry, 27(3), p.431 - 441, 2003/07
Times Cited Count:9 Percentile:45.16(Biology)One nanosecond molecular dynamics (MD) simulation was performed for two DNA segments each composed of 30 base pairs. The analysis of results was focused on the electrostatic energy that is supposed to be significant factor causing the disruption of DNA base stacking in DNA duplex and may also to serve as a signal toward the repair enzyme informing on the presence of the lesion. This electrostatic potential may signal presence of the lesion to the repair enzyme.
Pinak, M.; Laaksonen, A.
Molecular Mechanisms for Radiation-Induced Cellular Response and Cancer Development, p.266 - 273, 2003/00
The molecular dynamics (MD) simulations of DNA mutagenic oxidative lesion - 7,8-dihydro-8-oxoguanine (8-oxoG), single and complexed with the repair enzyme - human oxoguanine glycosylase 1 (hOGG1), were performed for 1 nanosecond (ns) in order to determine structural changes at the DNA molecule and to describe a dynamical process of DNA-enzyme complex formation. The broken hydrogen bonds resulting in locally collapsed B-DNA structure were observed at the lesion site. In addition the adenine on the complementary strand (separated from 8-oxoG by 1 base pair) was flipped-out of the DNA double helix. In the case of simulation of DNA and repair enzyme hOGG1, the DNA-enzyme complex was formed after 500 picoseconds of MD that lasted stable until the simulation was terminated at 1 ns.
Pinak, M.
JAERI-Research 2002-016, 31 Pages, 2002/09
The molecular dynamics (MD) simulations of DNA mutagenic oxidative lesion 8-oxoG, single and complexed with the repair enzyme hOGG1, were performed. In the case of simulation of single DNA molecule the broken hydrogen bonds resulting in locally collapsed B-DNA structure were observed at the lesion site. In addition the adenine on the complementary strand was flipped-out of the DNA double helix. In the case of simulation of DNA and repair enzyme hOGG1, the DNA-enzyme complex was formed after 500 picoseconds of MD that lasted stable until the simulation was terminated at 1 ns. The N-terminus of arginine 313 was located close to the phosphodiester bond of nucleotide with 8-oxoG enabling chemical reactions between amino acid and lesion. Phosphodiester bond at C5'of 8-oxoG was at the position close to the N-terminus of arginine 313. The water mediated hydrogen bonds network was formed in each contact area between DNA and enzyme further enhancing the stability of complex.
Ishida, Hisashi
Journal of Biomolecular Structure and Dynamics, 19(5), p.839 - 851, 2002/05
Times Cited Count:11 Percentile:26.27(Biochemistry & Molecular Biology)no abstracts in English
Ishida, Hisashi
Proceedings of the International Conference on Bioinformatics 2002 (CD-ROM), 6 Pages, 2002/02
no abstracts in English
JAERI-M 94-070, 97 Pages, 1994/03
no abstracts in English